He Wang, Xiaoyu Zhang, Dongdong Wang, Qianqian Jiang, Yue Sun, Baofeng Zhao, Zhen Liang, Guangyan Qing*, Bo Jiang*, Lihua Zhang and Yukui Zhang
Chem. Sci., 2025, DOI: 10.1039/D5SC01557J
https://doi.org/10.1039/D5SC01557J
Recently, our group, in collaboration with the research team led by Researcher Zhang Lihua and Associate Researcher Jiang Bo from the Biomolecular Efficient Separation and Characterization Research Group (Group 1810), has made significant progress in the study of protein N-phosphorylation modification. Based on phage display technology, the research team screened and obtained the first affinity peptide ligand that could specifically recognize the N-PO₃ group, and further developed the first N-phosphopeptide enrichment material involving inorganic solvents and metal ions, achieving efficient enrichment and in-depth analysis of the atypical N-phosphorylated proteome.
N-phosphorylation is an important but rarely studied post-translational modification, which is widely present in life processes such as histone modification, signal transduction, and metabolic regulation. Due to the chemical instability of the N-P bond, its modification sites are highly prone to hydrolysis. For a long time, there has been a lack of effective enrichment strategies, which has seriously hindered the systematic analysis of its biological functions. To overcome this challenge, the collaborative team innovatively utilized phage display technology to precisely screen out peptide ligands that could efficiently recognize N-phosphopeptide. These ligands were then covalently modified and fixed on the surface of magnetic agarose materials, establishing a highly selective and reproducible enrichment system. Under neutral conditions, the efficient enrichment and mass spectrometry detection of N-phosphopeptide could be accomplished.
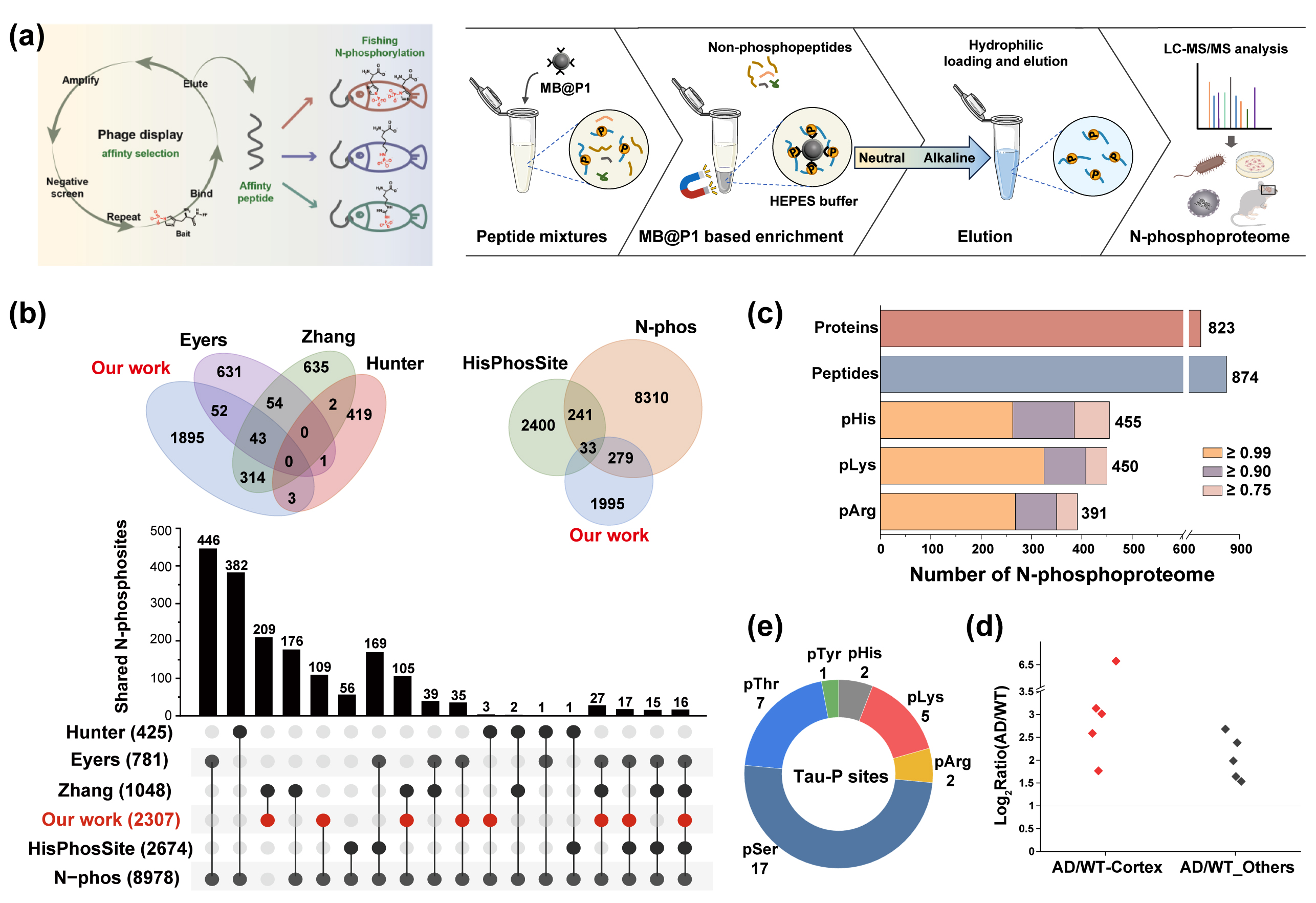
Through this strategy, the research team successfully depicted the panoramic map of atypical N-phosphorylation modifications in a variety of complex samples. A total of 2,876 N-phosphorylation sites (including pHis, pLys and pArg) were identified in HeLa cells, which was 2 to 5 times the depth of the existing data. 308 sites were also captured in Escherichia coli, significantly enhancing the detection throughput of modifications in microbial samples. It is particularly worth mentioning that the research team has for the first time systematically depicted the N-phosphate modification network at the nuclear subcellular omics level, revealing its important functions in key biological processes such as DNA repair, chromatin remodeling, and transcriptional regulation. Meanwhile, the study also applied this strategy to the brain tissues of Alzheimer's disease (AD) mouse models and found that the N-phosphate modifications of multiple Tau proteins and neurocytoskeletal related proteins were significantly upregulated, suggesting that this modification is closely related to the pathogenesis of AD and providing new biomarker clues for the research of neurodegenerative diseases. This work is currently the first in the world to propose and achieve an N-phosphorylation enrichment strategy without metal ions and organic solvents, providing a key tool for the systematic study of N-phosphorylation modification. At the same time, it offers a new solution for the highly challenging low-stability modification in PTM research.
The related achievement was published in Chemical Science under the title "Affinity peptide ligands: New tools for chasing non-canonical N-phosphoproteome". The first authors of this work are Wang He, a doctoral student from Group 1810, and Zhang Xiaoyu, an assistant researcher in our group. The above-mentioned research work was supported by the National Natural Science Foundation of China, the Innovation Fund of our institute and other projects. (Written and photographed by Zhang Xiaoyu)