Deciphering endogenous SUMO-1 landscape
Time:2024-12-30 15:55 Author:Xiaoyu Zhang
Xiaoyu Zhang, Bowen Zhong, Yue Sun, Dan Liu, Xiancheng Zhang, Dongdong Wang, Cunli Wang, Huiling Gao, Manli Zhong, Haijuan Qin, Yang Chen, Zhiying Yang, Yan Li, Haijie Wei, Xindi Yang, Yukui Zhang, Bo Jiang,* Lihua Zhang,* and Guangyan Qing*
Chem. Sci., 2024, 10.1039/D4SC07379G
https://pubs.rsc.org/en/Content/ArticleLanding/2025/SC/D4SC07379G
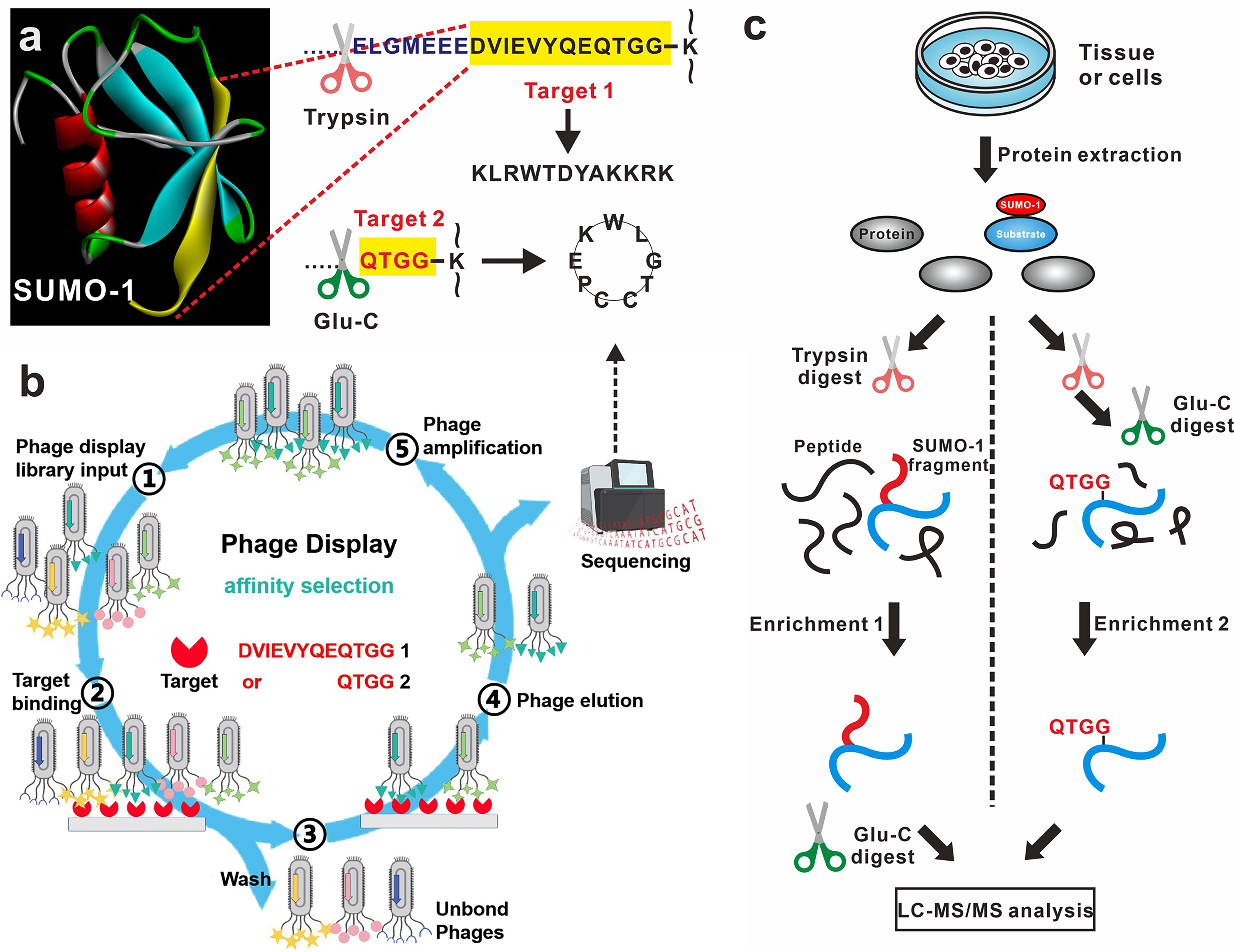
Small ubiquitin-like modifier (SUMO) plays a pivotal role in diverse cellular processes and is implicated in diseases such as cancer and neurodegenerative disorders. However, large-scale identification of endogenous SUMO-1 faces challenges due to limited enrichment methods and its lower abundance compared to SUMO-2/3. Here we propose a novel combinatorial peptide strategy, combined with anti-adhesive polymer development, to enrich endogenous SUMO-1 modified peptides, revealing a comprehensive SUMOylation landscape. Utilizing phage display, we successfully identified a linear 12-mer and a cystine-linked cyclic 7-mer peptide ligand, specifically designed to target the C-terminal regions of SUMO-1 remnants. Building upon their high affinities and satisfactory complementarity, we developed the first artificial SUMO-1 enrichment materials, ultimately established a combinatorial peptide strategy that facilitates a comprehensive analysis of endogenous SUMO-1 modified proteome in both cellular and tissue contexts. We successfully mapped 1312 SUMOylation sites in HeLa cells and 1365 along with 991 endogenous SUMOylation proteins in Alzheimer's disease (AD) mouse brain tissues. Notably, our method uncovered a significant upregulation of SUMO-1 in AD mouse brain tissue, providing new insights into SUMOylation’s role in disease. Overall, this work represents the most thorough exploration of SUMO-1 modified proteomics and offers robust tools for elucidating the roles of SUMO-1's biological significance.