Highly Efficient Separation of Methylated Peptides Utilizing Selective Complexation between Lysine and 18-Crown-6
Qianying Sheng, Cunli Wang, Xiaopei Li, Hongqiang Qin, Mingliang Ye, Yuting Xiong, Xue Wang, Xiuling Li, Minbo Lan*, Junyan Li, Yanxiong Ke, Guangyan Qing* and Ximiao Liang
Anal. Chem., 2020, 92, 15663−15670.
DOI: 10.1021/acs.analchem.0c04158
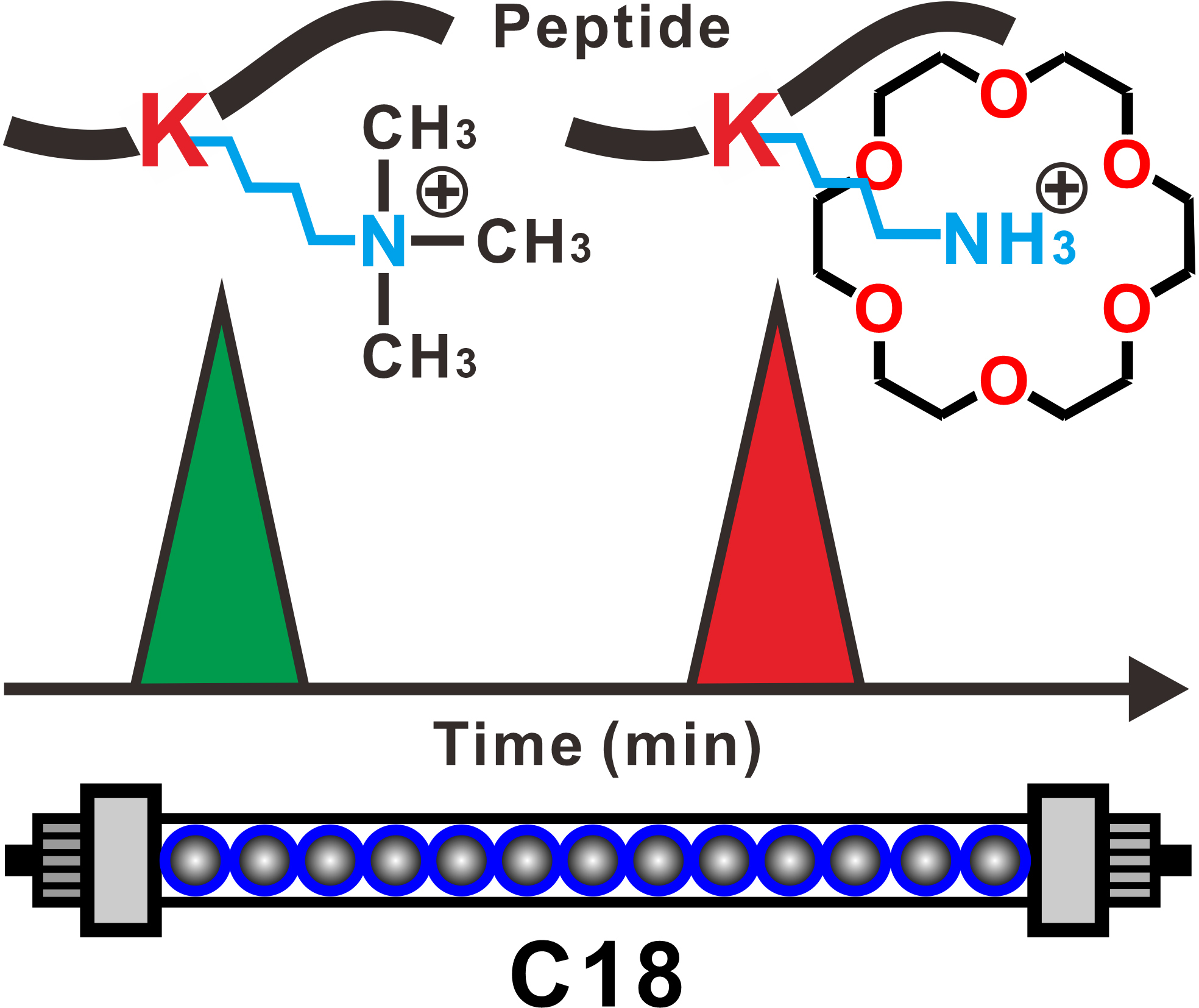
Protein methylation is one of the most common and important post-translational modifications and plays vital roles in epigenetic regulation, signal transduction and chromatin metabolism. However, due to diversity of methylation forms, slight difference between methylated sites and non-modified ones, and ultra-low abundance, it is extraordinarily challenging to capture and separate methylated peptides from biological samples. Here, we introduce a simple and highly efficient method to separate the methylated and non-methylated peptides based on using 18-crown-6 as a mobile phase additive in high-performance liquid chromatography (HPLC). Selective complexation between lysine and 18-crown-6 remarkably increases the retention of the peptides on a C18 stationary phase, leading to an excellent baseline separation between the lysine methylated and non-methylated peptides. A possible binding mechanism is verified by nuclear magnetic resonance titration, biolayer interferometry technology and quantum chemistry calculation. Through establishment of a simple enrichment methodology, a good selectivity is achieved and four methylated peptides with greatly improved signal-to-noise (S/N) ratio are successfully separated from a complex peptide sample containing 10 fold of bovine serum albumin (BSA) tryptic digests. By selecting rLys N as an enzyme to digest histone, methylation information in the histone could be well identified based on our enrichment method. This study will open an avenue and provide a novel insight for selective enrichment of lysine methylated peptides in post-translational modification proteomics.